Overview
The RH blood group system and molecular bases of D variants
The RH blood group system is the second most important system in transfusion medicine, after the ABO system. Recent reviews on this system include:
The two 10-exon genes of the RH system, named RHD and RHCE are located on chromosome 1 (1p34.3-1p36.1)
Each gene encodes a protein of 417 amino acids. The initial methionine is cleaved post-transcriptionally
The Figures below show the position of the RhD protein in the lipid bilayer, according to
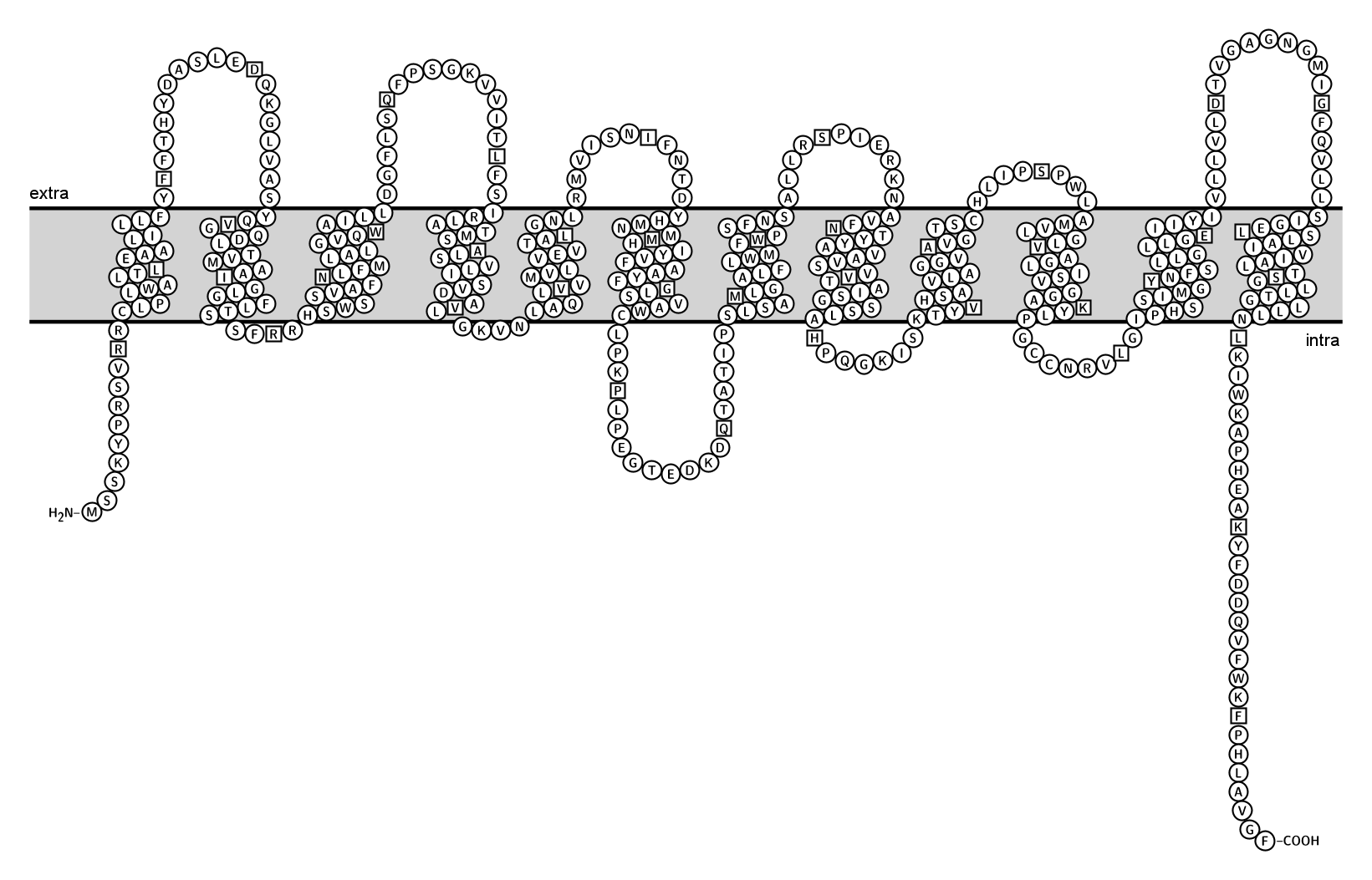
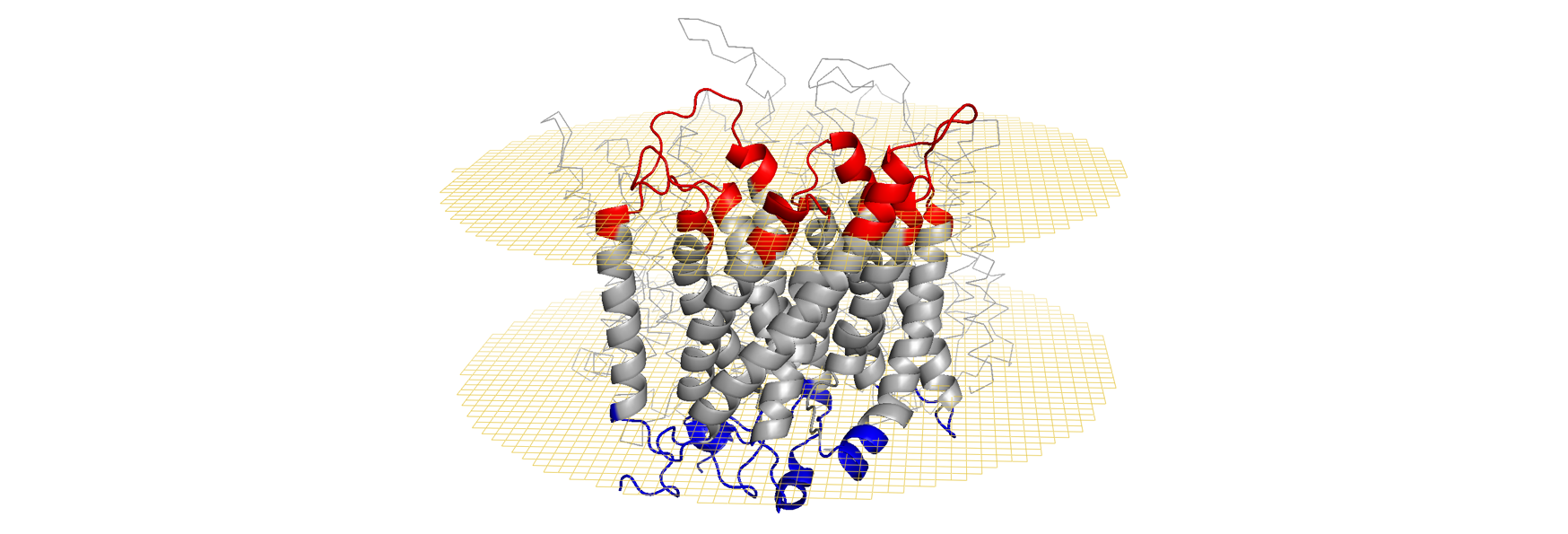
The reference sequences of RHD and RHCE in Genbank
Several nomenclatures are commonly used for the RH blood group system
Many genotyping methods exist
Many subtypes exist in some allele "clusters"
- DAU alleles
12070041 ,27480171 , - DIV alleles
23461862 , - DAR alleles
16584437 ,23902153 , - DVI alleles
9490704 , - DIII alleles
21745213 ,20088832 ,16584437 .
It can be interesting to organize RHD alleles into phylogenetic trees to reflect their potential relationships and how they arose.
Sources and data processing
Relevant articles are regularly searched in PubMed using requests with different keywords ("RHD alleles", "D variant", "RHD blood group variants" etc.).
Non-peer reviewed data proceed from:
- non-redundant informative data from Genbank entries
23193287 , - conference abstracts for some non-redundant data.
Each piece of data is associated an entry as described by the sources' authors. The allele assignment provided by the authors has not been reinterpreted. Please refer to the source material for methodology and allele assignment.
The sources and citations for all information are included so users can easily identify the source and access the methods and more details, as needed.
The data included in RHeference remain the responsibility and property of the sources' authors and the journals in which the data have been published.
How to browse and query the database
To easily access information, there are several possibilities, accessible from the header.
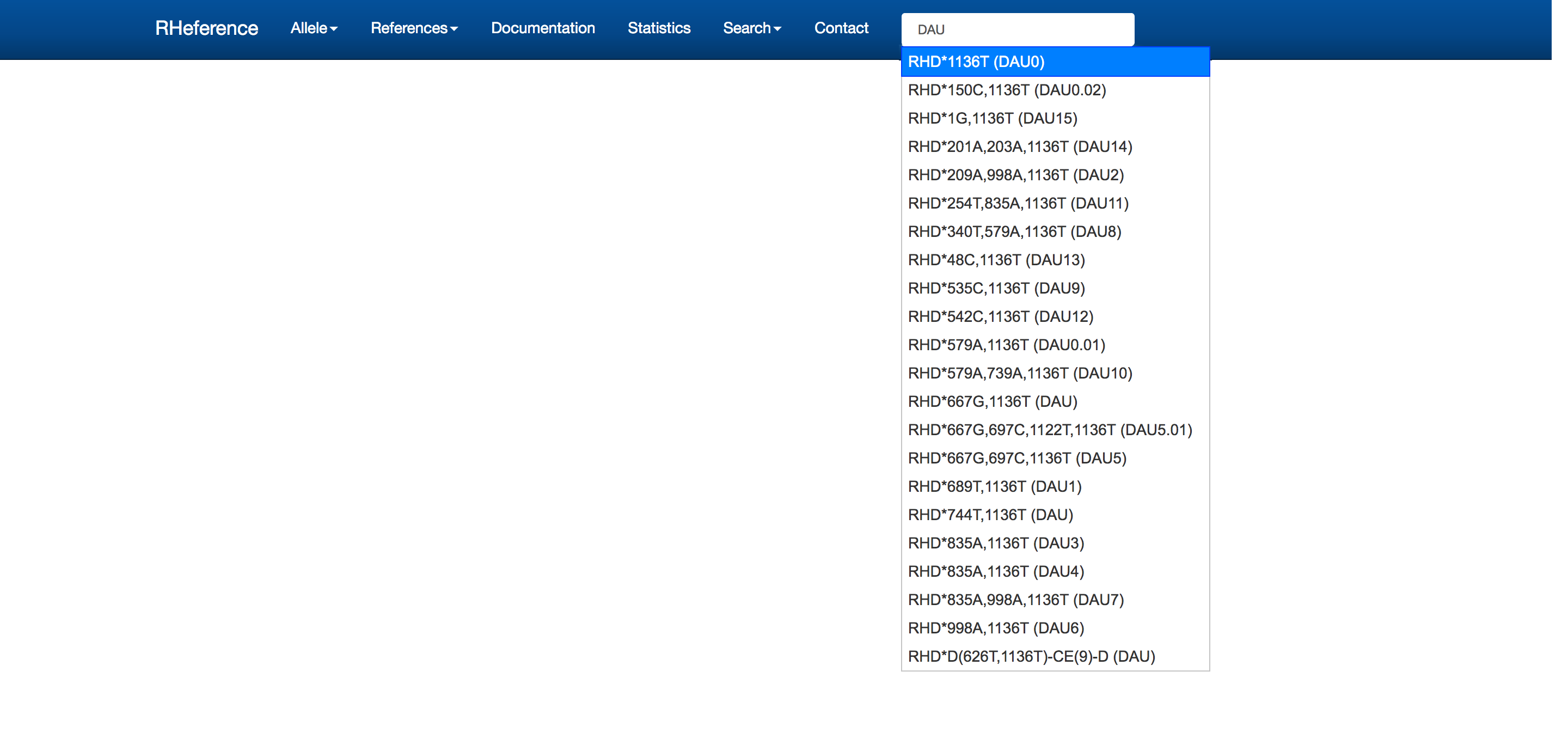
- A quick search box to the right: the query applies to allele names, nucleotide and amino acid positions. E.g. if the user types “DAU” in the box, RHeference entry names that match this text will appear. The user should directly click on the allele they wish to access and will be redirected to the allele’s page.
- From the drop-down menu “Search”, the page “By name” enables a simple query by keywords.
- From the drop-down menu “Search”, the page “By mutation” enables a query combining any number of nucleotide and/or amino acid positions.
- From the drop-down menu “Search”, the page “In exons” enables a search by nucleotide mutation(s) within the coding sequence. The user must click on each exon to reveal the list known of mutations within this exon. The user can select one or more mutations in one or more exons. With the restrictive option “All positions selected present”, the output will only show alleles combining all the mutations selected. With the option “Any position selected present”, the output will show alleles containing one or more of the mutations selected.
- From the drop-down menu “Search”, the page “Complex” enables elaborate queries with advanced and/or combined criteria.

Moreover, RHeference allows easy, “horizontal” navigation through dedicated links. By clicking on any characteristic in an allele or entry (a mutation, a phenotype, an article, etc.), the user is redirected to a list of all the entries sharing the same characteristic. The example below shows the output when the user clicks on the mutation c.809T>G from within any allele including this genetic variant.

Acknowledgements
The authors would like to thank all the people who have shown their enthusiastic support for this project.
Example
Each entry in RHeference contains subsections. By default, the detailed data is not displayed, but each subsection can be expanded with the arrows and plus signs on the right. The entry for the RHD*38 (RHD*DNT) allele with some of the subsections in its expanded version is used below to detail each subsection. Clicking on any characteristic of an allele or entry (e.g. a mutation, a phenotype, etc.) underlined in blue redirects the user to a list of all the RHeference entries that share this characteristic.
Names and ISBT classification

The header of each entry is the official ISBT nomenclature and the ISBT table in which it can be found, if applicable.
More names
Other names can be found in this section.
Molecular data
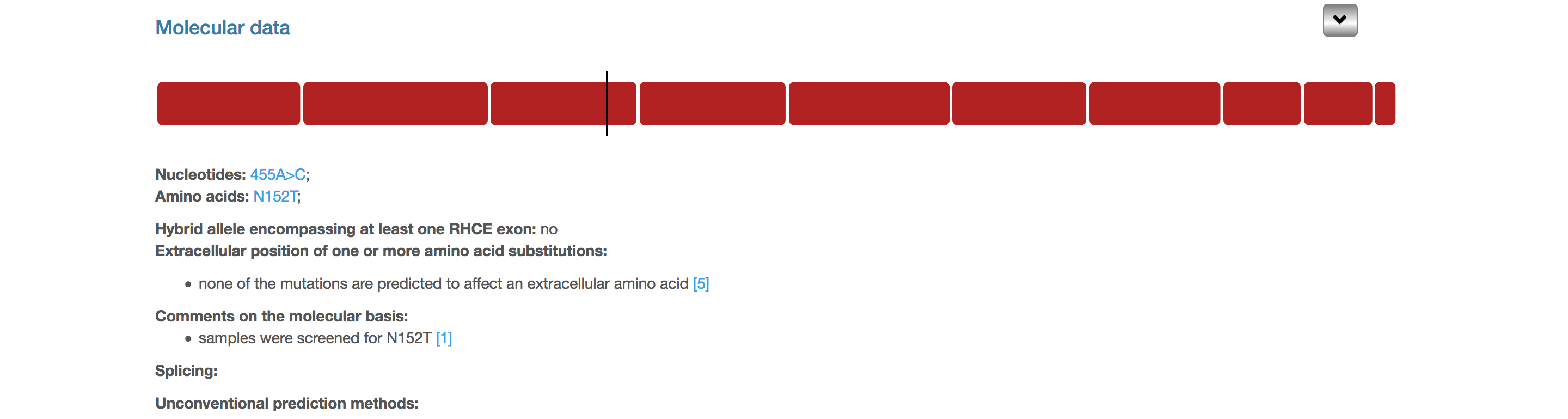
This section shows genetic variations found in the allele (nucleotide and amino acid) if applicable, whether the allele is a hybrid between RHD and RHCE, relevant information about the molecular analyses performed in the studies describing this allele, where the substitutions are predicted to be relative to the lipid bilayer, and how the variations affect or are predicted to affect splicing and the protein structure.
In the compact view, only the schema of the allele is shown in this section, with RHD exons in red, exons derived from RHCE in blue, deleted exons in white with a dotted contour and the mutations as vertical black lines. Hovering over the exons and the mutations reveals pop-ups with nucleotide and amino acid positions (black box in the screenshot).
More molecular data
This section lists comments regarding the molecular bases, the genotyping methods, hybrid alleles, splicing, the position of the amino acid substitutions relative to the lipid bilayer, etc.
Phenotype
This section contains phenotypic data for the allele. The main phenotype (e.g. “positive” in this example) is an expert annotation, deduced from the available published data and visible even when the subsection is collapsed. The most recent update of this expert annotation is shown. Expanding the section with the arrow to the right reveals more information.

Reports by D phenotype
In this section, the reported D phenotypes are gathered and organized into 7 categories:
- D positive
- undetailed ambiguous D phenotype
- discrepant D phenotype: D-negative or D-positive depending on anti-D reagents and techniques within the same report
- weak D phenotype
- very weak D phenotype, which may be missed depending on techniques and reagents
- DEL phenotype refers to a very weak D positive phenotype only detectable by adsorption-elution methods.
32203009 DEL alleles may appear D negative (RH:—1) if no fixation-elution has been performed.16181205 ,29043831 . - D negative
For some RHCE alleles, anti-D or anti-D-like reactivity has been observed, including sometimes in the absence of RhD-specific amino acids.
Phenotyping result may differ significantly for the same allele depending on samples and phenotyping methods or reagents (e.g. some variants may type negative or weakly positive in different studies, depending on whether direct or indirect agglutination testing was performed)
Other RH phenotypes
Many samples have been tested for RH antigens other than D.
When the absence of C, E, c or e antigen expression has been inferred from the haplotypes with which the RHD allele has been described, it is mentioned as a comment. E.g. theRHD*DNT allele has consistently been described with an RH:-2,-3,4,5 phenotype, and it is reasonable to assume that the allele doet not express neither C nor E.
For serologic and/or molecular descriptions of the different RH antigens,
- RH2 (C), RH3 (E), RH4 (c), RH5 (e):
7702587 ,7513151 ; - RH6 (ce), RH7 (Ce), RH22 (CE), RH27 (cE) are compound antigens ;
- RH8 (CW), RH9 (CX):
7620172 ; - RH10 (V), RH20 (VS):
13251871 ,9767746 ,9024488 ,23286557 ,19040491 ,13499241 ; - RH11 (EW):
14996199 ,13239995 ; - RH12 (G):
804766 ,8669081 ; - RH17 (Hr0):
15318861 ,14764967 ,21517889 ,10807539 ; - RH18 (Hr), RH19 (hrS):
8036793 : - RH23 (DW):
13879234 ,14154107 ,17900276 ,12076297 ; - RH26 (c-like):
9426634 ,14242756 ,8023390 ,17002624 , antithetical to RH55; - RH29:
9218155 ,25413218 ; - RH30 (Goa):
21729099 ,4971757 ,4965288 ,23461862 ; - RH31 (hrB):
23286557 ,19624490 ,19040491 ; - RH32:
2511647 ,9316225 ,2510749 (in French), antithetical to RH46; - RH33 (R0Har):
1926324 ,8614957 ,2455369 ,21729099 ,3095959 ; - RH34 (HrB):
1763497 ,23286557 ,19624490 ,19040491 ; - RH36 (Bea):
19951310 ; - RH37 (Evans):
414453 ,8808597 ,10729811 ; - RH40 (Tar):
116544 ,6418437 ; - RH43 (Crawford):
16934069 ,20609196 , antithetical to RH58; - RH44 (Nou):
6784228 ; - RH45 (Riv):
21729099 ; - RH46 (Sec):
2511647 ,2510749 (in French), antithetical to RH32; - RH47 (Dav):
6808647 ; - RH48 (JAL):
21477150 ,19170983 ,19192256 ,2118698 ,2118699 ,19207167 ,20233350 , antithetical to RH57; - RH49 (STEM):
22738288 ,19192256 ,21477150 ,8038895 ; - RH50 (FPTT):
7519797 ,21729099 ; - RH51 (MAR):
8079453 ; - RH52 (BARC):
16584438 ; - RH53 (JAHK):
16078918 ,20233350 ; - RH54 (DAK):
21477150 ; - RH55 (LOCR):
17002624 , antithetical to RH26; - RH56 (CENR):
15225246 ; - RH57 (CEST):
19192256 , antithetical to RH48; - RH58 (CELO):
20609196 , antithetical to RH43 - RH59 (CEAG):
26173592 ; - RH60 (PARG):
28144953 ; - RH61:
23772606 ; - RH62 (CEWA):
30421425 .
Serology with monoclonal anti-D
The D antigen is considered to be composed of a mosaic of epitopes. By testing D variants with monoclonal antibodies and cross-matching, epitope patterns have been proposed
Short summaries of the published findings are reported here, pincluding where published epitope patterns can be found. For the list of anti-D tested and detailed reactivities, see the linked publications.
Antigen Density (Ag/RBC)
D antigen density is the number of D antigens per RBC
As all other data, D antigen density is entered in RHeference as published in the source material. Methods vary widely between studies: one or more samples are tested, one or several monoclonal or polyclonal anti-D reagents are used, different control samples are selected... Some studies report results as values and standard deviations, others as medians, ranges, quartiles… Because of this heterogeneity, we recommend referring to the Materials and Methods section of the source articles for detailed information.
More phenotype data
Some other phenotype data are listed here, including the Rhesus similarity index which has been occasionally used to assess how similar D variants are to the standard D antigen
Haplotype
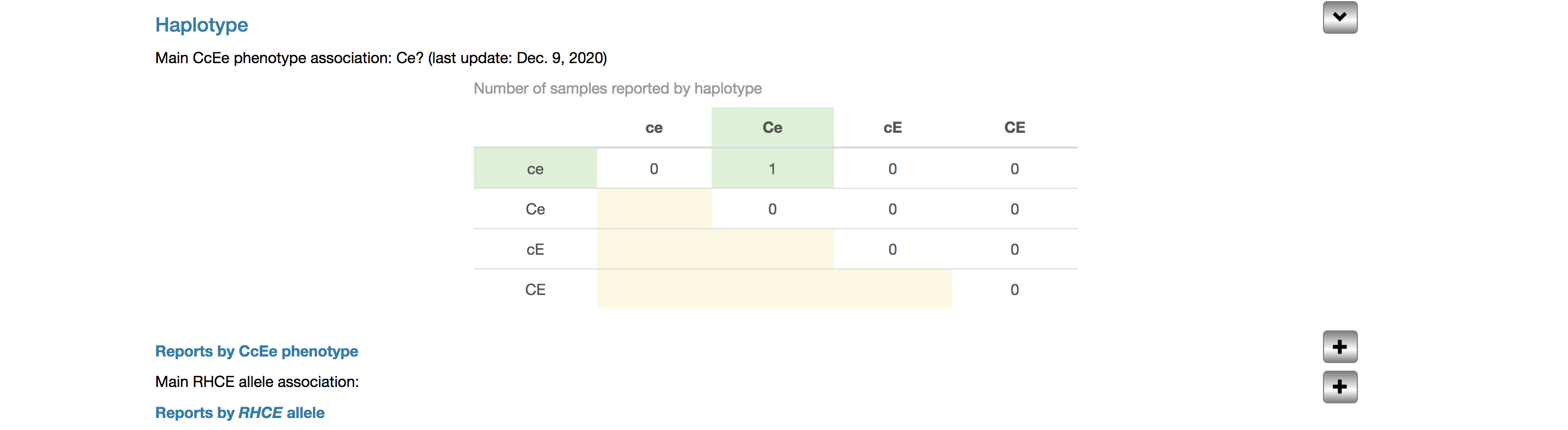
This section is dedicated to the relationship between RHD and RHCE alleles, and by extension, phenotypes.
Due to the proximity of RHD and RHCE genes on chromosome 1, specific RHD alleles are frequently linked with specific RHCE alleles to form haplotypes. Knowing RHCE phenotypes can help target analyses in favor of the most likely D variants.
For some alleles and phenotypes, the association is frequent and well known, or several associations are possible. For others, the descriptions with each phenotype and RHCE alleles can be informative to deduce the most likely haplotype association.
Main CcEe phenotype association
The main (or most likely) CcEe phenotype with which the allele has been associated is an expert annotation, deduced from the available published data and visible even when the subsection is collapsed. The most recent update of this expert annotation is shown Expanding the section with the arrow to the right reveals more information.
Reports by CcEe phenotype
All CcEe phenotypes reported are organized in the table (e.g. RHD*DNT has been reported in a Ccee sample), to underline the haplotype associations, and listed as text below it.
Main allele association and reports by RHCE allele
Recent allele descriptions and population studies report molecular typing of the whole haplotype. Like above, the main (or most likely) allele association (e.g. RHCE*01.01 for RHD*10.04) is an expert annotation, deduced from the available published data. The most recent update of this expert annotation is shown. The detailed data are visible when the subsection is expanded.
Alloimmunization

This section contains data regarding antibody formation: in individuals (“carriers”) expressing the variant (who received red blood cells positive for the standard antigen), and in recipients negative for the antigen (who received red blood cells from a donor expressing the variant).
The summaries of these data are expert annotations, deduced from the available published data. The most recent update of the expert annotation is shown. The detailed data are visible when the subsection is expanded.
Antibodies in carriers
This section contains data regarding antibody formation in carriers of each variant.
Both allo- and auto-antibodies have been listed, as well as antibodies not specified to be one or the other. Whenever it is available, pertinent serologic data is included. Arguments in favor of an allo-antibody include: negative autologous control, negative direct antiglobulin test (DAT), negative elution results (i.e. no anti-D in the eluate), negative auto-adsorption (i.e. the antibody could not be auto-adsorbed)
In many studies, the information is only partly detailed. Thus, “ND” stands for: “not done” or “not disclosed” (implied: in the study), as the difference is not always obvious. Antibodies are often simply listed as alloantibodies with no additional data. We have kept the authors’ assignment but recommend users to be cautious and always read the original source material.
We have chosen to include anti-D presented in conference abstracts in RHeference. It is sometimes argued that the existence of anti-D which have not made it into peer-reviewed journals may be doubted
Antibodies in recipients
This section is dedicated to anti-D formation in D negative recipients, when the D variant carrier is the blood donor. Indeed, some DEL and very weak D phenotypes have been shown to induce anti-D reactivation or formation in recipients
Reports

This section details the situations, and ethnicities and populations in which the allele has been described. The summary of these reports a curated summary of the reports is shown. It is an expert annotation deduced from the available published data, visible even when the subsection is collapsed. The most recent update of this expert annotation is shown. All the situations and ethnicities in which the allele has been reported are visible when the subsection is expanded.
The genetic diversity of the human species is responsible for a non-homogenous distribution of alleles worldwide, including alleles of blood group genes.
Most studies rely on an initial phenotyping step to identify samples with specific serologic features (weak D phenotype in most cases, or D negative phenotype in blood donors) before proceeding to genotyping. Any prevalence estimated from these works are biased, especially for RHD alleles associated with quasi-normal D antigen expression. This section does not aim to establish true prevalences in different populations, to give users a summary overview of the literature and help them target alleles of interest.
Consequently, this section gathers data regarding the number of samples, brief summaries of the contexts in which each allele has been reported, and ethnicities or locations. When studies do not mention the probands’ ethnicities, we list the population (e.g. "in the French population") or if it could not be inferred, the laboratory's localization (e.g. "reported by a German lab").
Links

In this section, the urls to other websites for the allele can be accessed (if applicable):
- Genbank accession number(s)
23193287 , - link(s) to The Human RhesusBase
25538538 , - a link to Erythrogene, performing a search with the mutation(s) of the allele.
29296939 . If there are no matches in Erythrogene, this search returns no results.
References
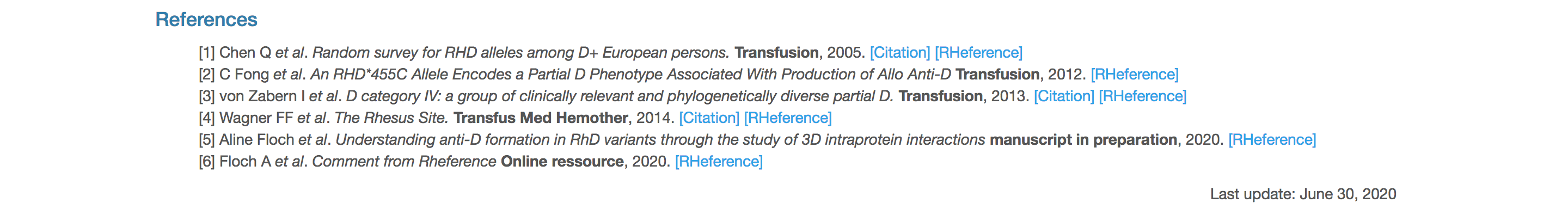
All the sources of the data presented for the entry are listed in this section and numbered. Clicking on any numbered reference on the page jumps down to and highlights the corresponding footnote.
Click the word [Citation] next to each source to be redirected to either: the Digital Object Identifier when possible or the Pubmed entry (by default). Click [RHeference] to be redirected to a dedicated page within the RHeference database tracing all allele annotations for this source.
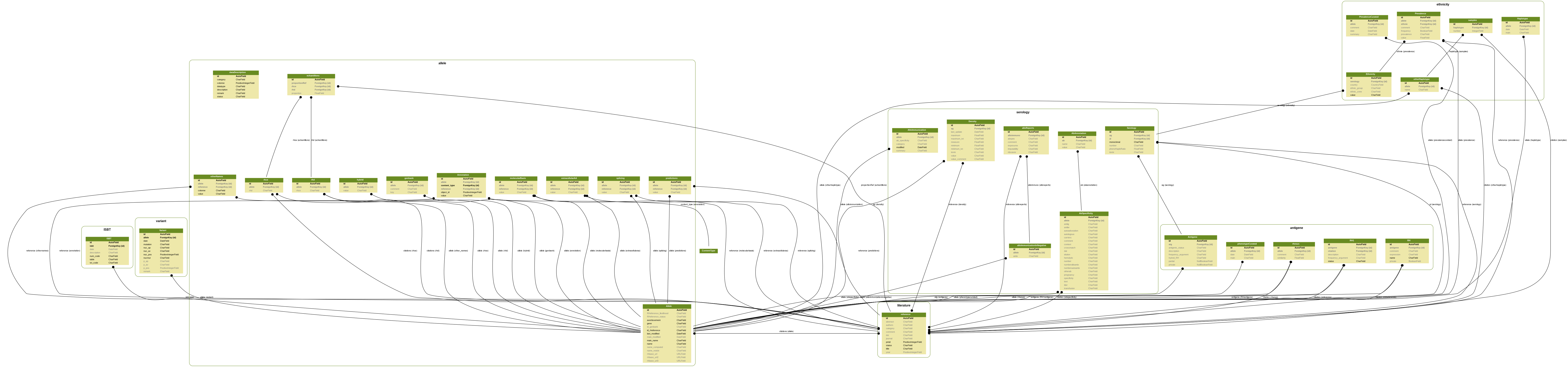
Database
RHeference is based on BioDjango, an open Python framework for bioinformatics. The schema below shows the structure of the database, with the complex relationships between data.
Funding and licensing
This study was supported by grants (2016 and 2018) from Laboratory of Excellence GR-Ex, reference ANR-11-LABX-0051. The labex GR-Ex is funded by the program "Investissements d’avenir" of the French National Research Agency, reference ANR-11-IDEX-0005-02.
This work was performed using high performing computing resources from GENCI-CINES (Grants 2018-A0040710370, 2020- A0070710961 and 2020-A0080711465).

The database is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License.
Collaborators
Data collected and curated by
Aline Floch, Christophe Tournamille, France Pirenne, Etablissement français du sang Ile de France, Université Paris Est Creteil, INSERM, IMRB, Creteil, France
Site created by
Stéphane Téletchéa, Unité Fonctionnalité et Ingénierie des Protéines, Team Conception de Protéines in silico, University of Nantes, France
Site hosted by
Alexandre G. de Brevern, Dynamics of Structures and Interactions of Biological Macromolecules, Biologie Intégrée du Globule Rouge UMR_S1134, Inserm, Univ. Paris, Univ. de la Réunion, Univ. des Antilles, Institut National de la Transfusion Sanguine (INTS), Paris, France.
FAQ
Some allele entries seem incomplete.
This could be a bug, please tell us if you think this is the case. For most entries, though, there is simply no published data available for now. Please consider publishing your work to fill in the gaps, for the benefit of the whole scientific community!
Why is my article/this very interesting article/this allele not listed in your database?
We have performed an extensive review of the literature and identified hundreds of relevant works. It may be that your article is in our to-do list and will soon be incorporated, or that we are not yet aware of this specific publication. Don’t hesitate to contact us to mention it. We will do our best to consider and incorporate relevant publications.
Are you planning on expanding the scope of RHeference?
Indeed, we hope to expand RHeference, at least to RHCE alleles. We are not sure yet whether we will consider other blood group systems in the future. In the meantime you can visit the ISBT website and the National Center for Blood Group Genomics website.
Do you accept personal communications?
At the moment, we are not considering unpublished data which has not at least been presented in abstract form at a Transfusion or Hematology conference. However, feel free to contact us, as our policy may change in the future.
I would like to see feature abc added, is it possible?
Nothing is impossible, contact us and we will consider your suggestions for future updates of RHeference.